Re: [筆記] R 的設定-平行運算 (CPU)
分享另一個平行運算的package: snowfall
以下是一個例子 for cross validation
### Example: Cross validation for choosing
### the tuning parameter of ridge regression
## Data
data(longley)
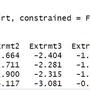
X <- as.matrix(longley[,1:5])
Y <- as.matrix(longley[,7])
## Normalization
norm <- function(a) (a-mean(a))/sd(a)
Y.N <- apply(Y, 2, norm)
X.N <- apply(X, 2, norm)
D <- cbind(Y.N, X.N)
n <- nrow(X.N)
p <- ncol(X.N)
## Ridge regression, given lambda
beta.R <- function(Y, X, nc, lambda) solve(t(X)%*%X +
lambda*diag(nc),t(X)%*%Y)
## cross validation, single CPU version
m <- 1001
lambda <- seq(0, 1, length=m)
CV <- sapply(1:m, function(j){
out <- sapply(1:n, function(i)
beta.hat <- beta.R(Y.N[-i,], X.N[-i,], p, lambda[j]))
out <- mean((Y.N - diag(X.N%*%out))^2)
})
plot(lambda, CV, type='l', xlab=expression(lambda), ylab='CV')
lambda.opt <- lambda[which(CV == min(CV))]
beta.opt <- beta.R(Y.N, X.N, p, lambda.opt)
## cross validation, parallel version
## in this case, the sample size is small but the number of lambda is large
## so we replace the outer loop by parallel computing
## prepare for parallel computing
library(snowfall) # parallel computing
sfInit(parallel = TRUE, cpus = 8, type="SOCK") # built four nodes
## Tell all nodes that following objects are going to be used
sfExport('Y.N', 'X.N','m','n','p','beta.R', 'lambda')
inner.loop <- function(j){
A <- apply(matrix(1:m,m,1), 1, function(i)
beta.hat <- beta.R(Y.N[-i], X.N[-i,], p, lambda[j]))
out <- mean((Y.N - diag(X.N%*%A))^2)
return(out)
}
CV <- sfSapply(seq(along=Y.N), inner.loop)
## evaluate the performance
wallTime <- function(expr) system.time(expr)[3]
wallTime(sapply(1:m, function(j){
out <- sapply(1:n, function(i)
beta.hat <- beta.R(Y.N[-i,], X.N[-i,], p, lambda[j]))
out <- mean((Y.N - diag(X.N%*%out))^2)
}))
wallTime(sfSapply(seq(along=Y.N), inner.loop))
sfStop()
--
※ 發信站: 批踢踢實業坊(ptt.cc)
◆ From: 218.164.73.129
推
06/26 19:38, , 1F
06/26 19:38, 1F
推
06/26 22:07, , 2F
06/26 22:07, 2F
討論串 (同標題文章)
完整討論串 (本文為第 2 之 2 篇):
R_Language 近期熱門文章
PTT數位生活區 即時熱門文章